Databricks#
You can install RAPIDS on Databricks in a few different ways:
Accelerate machine learning workflows in a single-node GPU notebook environment
Spark users can install RAPIDS Accelerator for Apache Spark 3.x on Databricks
Install Dask alongside Spark and then use libraries like
dask-cudffor multi-node workloads
Single-node GPU Notebook environment#
Create init-script#
To get started, you must first configure an initialization script to install RAPIDS libraries and all other dependencies for your project.
Databricks recommends using cluster-scoped init scripts stored in the workspace files.
Navigate to the top-left Workspace tab and click on your Home directory then select Add > File from the menu. Create an init.sh script with contents:
#!/bin/bash
set -e
# Install RAPIDS libraries
pip install --extra-index-url=https://pypi.nvidia.com \
"cudf-cu11" \
"cuml-cu11" \
"dask-cudf-cu11" \
"dask-cuda==24.06"
Launch cluster#
To get started, navigate to the All Purpose Compute tab of the Compute section in Databricks and select Create Compute. Name your cluster and choose “Single node”.
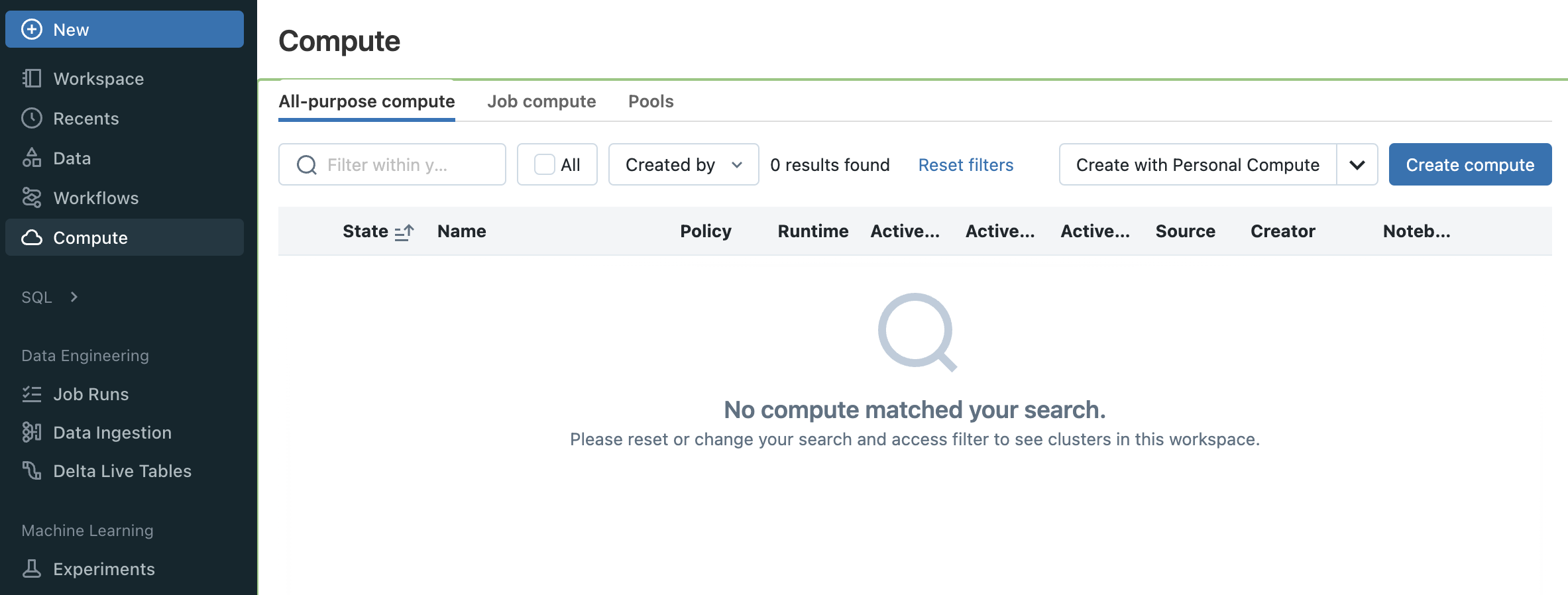
In order to launch a GPU node uncheck Use Photon Acceleration and select 14.2 ML (GPU, Scala 2.12, Spark 3.5.0) runtime version.
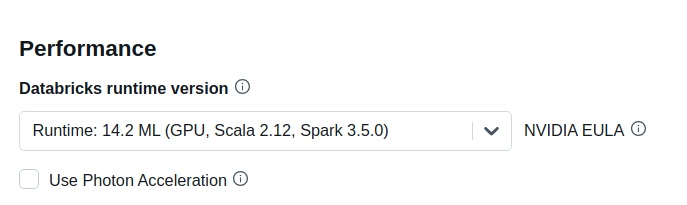
The “GPU accelerated” nodes should now be available in the Node type dropdown.
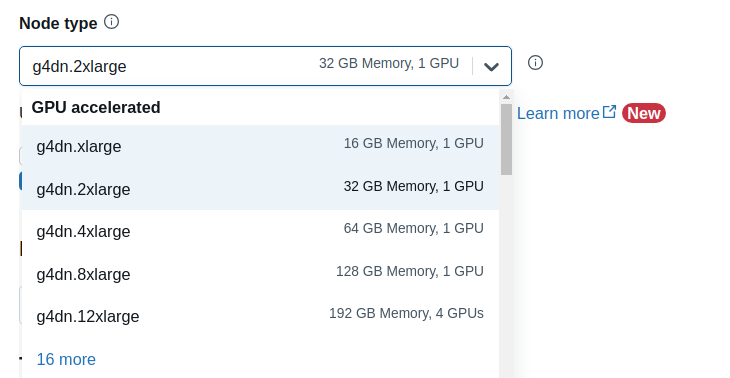
Then expand the Advanced Options section, open the Init Scripts tab and add the file path to the init-script in your Workspace directory starting with /Users/<user-name>/<script-name>.sh.
Select Create Compute
Test RAPIDS#
Once your cluster has started, you can create a new notebook or open an existing one from the /Workspace directory then attach it to your running cluster.
import cudf
gdf = cudf.DataFrame({"a":[1,2,3],"b":[4,5,6]})
gdf
a b
0 1 4
1 2 5
2 3 6
Quickstart with cuDF Pandas#
RAPIDS recently introduced cuDF’s pandas accelerator mode to accelerate existing pandas workflows with zero changes to code.
Using cudf.pandas in Databricks on a single-node can offer significant performance improvements over traditional pandas when dealing with large datasets; operations are optimized to run on the GPU (cuDF) whenever possible, seamlessly falling back to the CPU (pandas) when necessary, with synchronization happening in the background.
Below is a quick example how to load the cudf.pandas extension in a Jupyter notebook:
%load_ext cudf.pandas
%%time
import pandas as pd
df = pd.read_parquet(
"nyc_parking_violations_2022.parquet",
columns=["Registration State", "Violation Description", "Vehicle Body Type", "Issue Date", "Summons Number"]
)
(df[["Registration State", "Violation Description"]]
.value_counts()
.groupby("Registration State")
.head(1)
.sort_index()
.reset_index()
)
Upload the 10 Minutes to RAPIDS cuDF Pandas notebook in your single-node Databricks cluster and run through the cells.
NOTE: cuDF pandas is open beta and under active development. You can learn more through the documentation and the release blog.
Multi-node Dask cluster#
Dask now has a dask-databricks CLI tool (via conda and pip) to simplify the Dask cluster startup process within Databricks.
Install RAPIDS and Dask#
Create the init script below to install dask, dask-databricks RAPIDS libraries and all other dependencies for your project.
#!/bin/bash
set -e
# The Databricks Python directory isn't on the path in
# databricksruntime/gpu-tensorflow:cuda11.8 for some reason
export PATH="/databricks/python/bin:$PATH"
# Install RAPIDS (cudf & dask-cudf) and dask-databricks
/databricks/python/bin/pip install --extra-index-url=https://pypi.nvidia.com \
cudf-cu11 \
dask[complete] \
dask-cudf-cu11 \
dask-cuda==24.06 \
dask-databricks
# Start the Dask cluster with CUDA workers
dask databricks run --cuda
Note: By default, the dask databricks run command will launch a dask scheduler in the driver node and standard workers on remaining nodes.
To launch a dask cluster with GPU workers, you must parse in --cuda flag option.
Launch Dask cluster#
Once your script is ready, follow the instructions to launch a Multi-node cluster.
Be sure to select 14.2 (Scala 2.12, Spark 3.5.0) Standard Runtime as shown below.
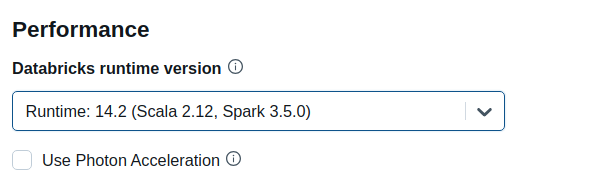
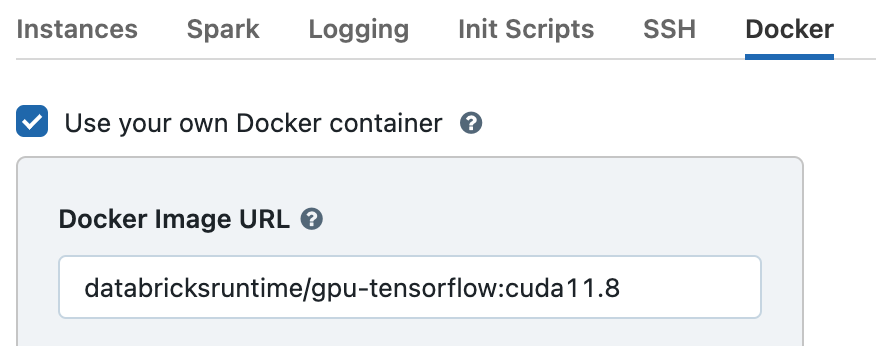
Then expand the Advanced Options section and open the Docker tab. Select Use your own Docker container and enter the image databricksruntime/gpu-tensorflow:cuda11.8 or databricksruntime/gpu-pytorch:cuda11.8.
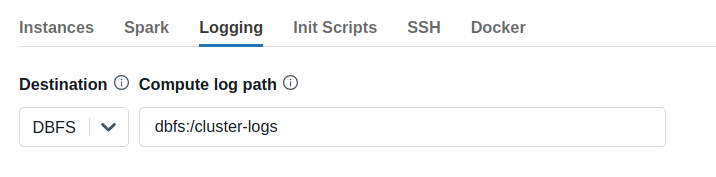
Configure the path to your init script in the Init Scripts tab. Optionally, you can also configure cluster log delivery in the Logging tab, which will write the init script logs to DBFS in a subdirectory called dbfs:/cluster-logs/<cluster-id>/init_scripts/.
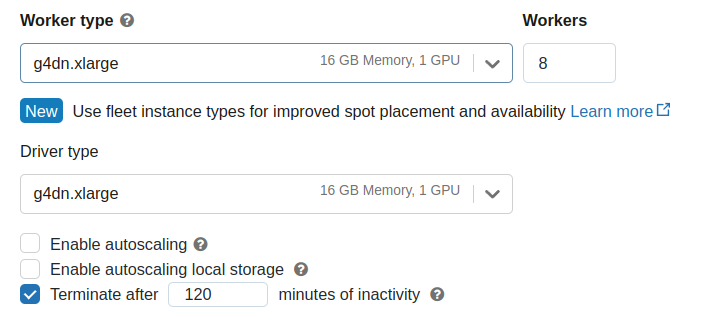
Once you have completed, you should be able to select a “GPU-Accelerated” instance for both Worker and Driver nodes.
Select Create Compute
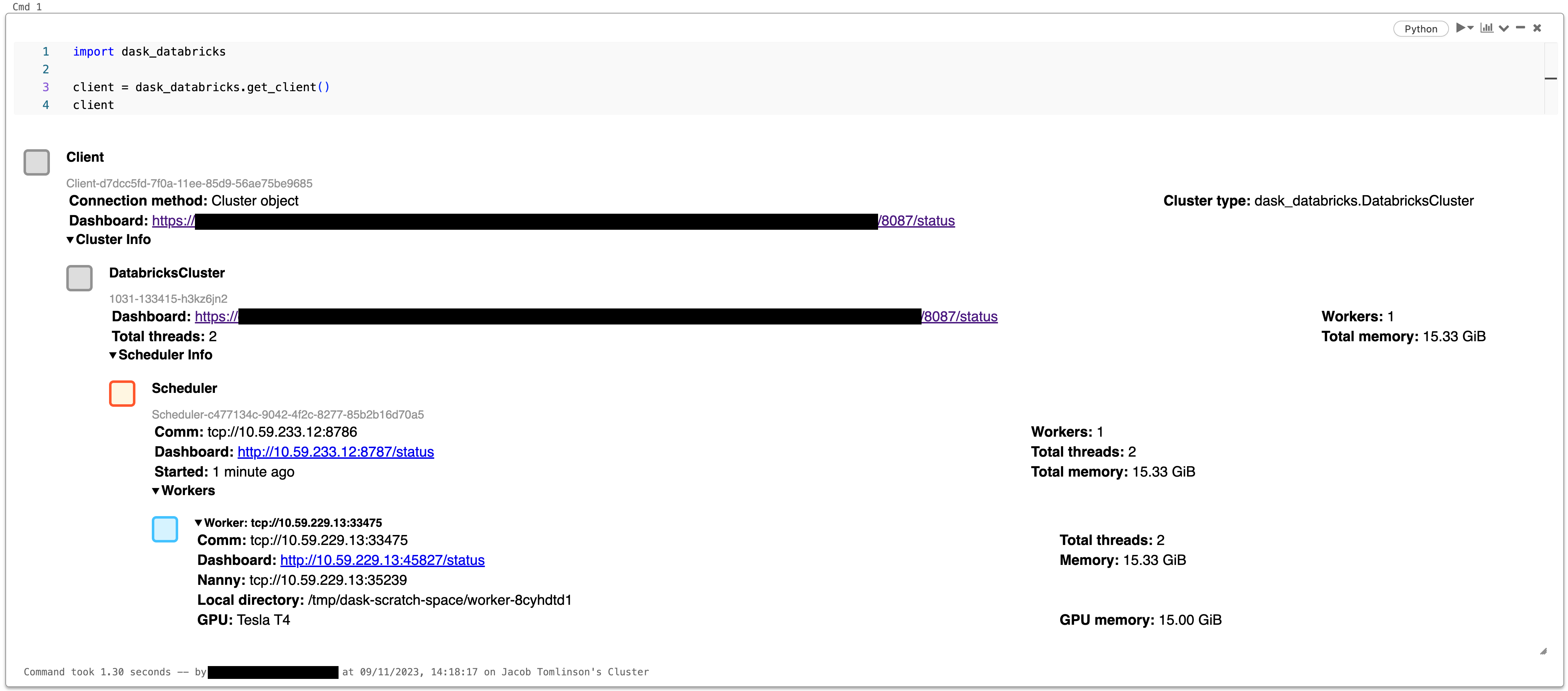
Connect to Client#
import dask_databricks
client = dask_databricks.get_client()
client
Dashboard#
The Dask dashboard provides a web-based UI with visualizations and real-time information about the Dask cluster status i.e task progress, resource utilization, etc.
The dashboard server will start up automatically when the scheduler is created, and is hosted on port 8087 by default. To access follow the URL to the dashboard status endpoint within Databricks.
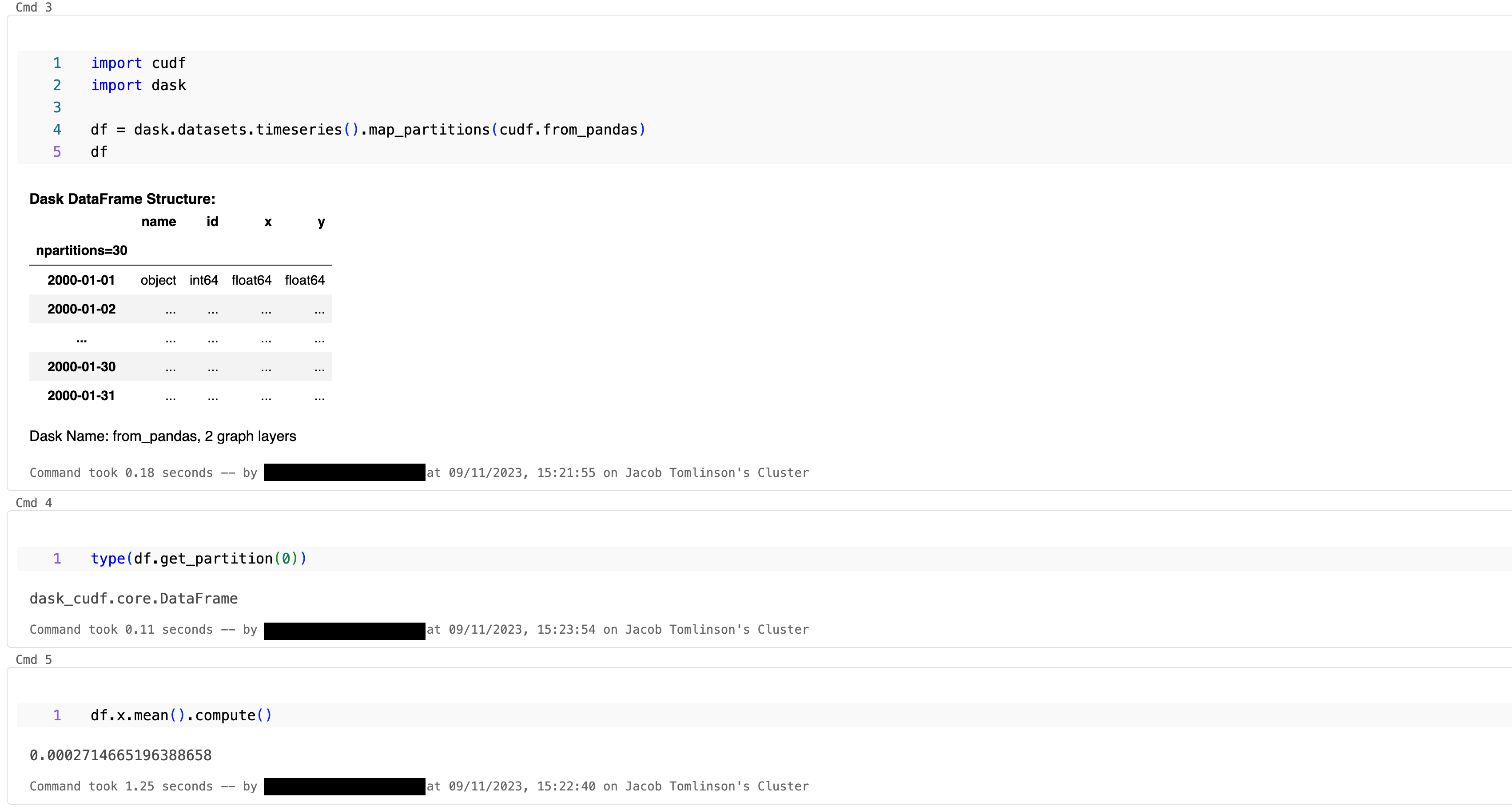
Submit tasks#
import cudf
import dask
df = dask.datasets.timeseries().map_partitions(cudf.from_pandas)
df.x.mean().compute()
Dask-Databricks example#
In your multi-node cluster, upload the Training XGBoost with Dask RAPIDS in Databricks example in Jupyter notebook and run through the cells.
Clean up#
client.close()
